One biomedical engineer’s frustration becomes another’s basis of success
UF Engineers solve 50+-year-old enzyme dissipation and distortion puzzle, opening the door to enzyme therapy for osteoarthritis, periodontal disease and, potentially, cancer
A 50-year-old problem
Since the 1960s, scientists have known that employing enzymes as drugs would afford new opportunities to treat diseases such as cancer, metabolic disorders, and autoimmunity. The body’s own ability to fight infection, enhance wound healing, and promote tissue regeneration are also tied to enzymes. To realize their therapeutic potential, though, approaches were needed to introduce enzymes at the right place in the body for a useful duration of time.
The biggest puzzle had been how to localize the enzyme at the site of activity – for example, inflammation in an arthritic knee – while avoiding widespread distribution that could result in off-target reactions throughout the patient’s body. For more than 50 years, the solution to that puzzle eluded researchers. Then, one day Benjamin Keselowsky, Ph.D., Professor at UF’s J. Crayton Pruitt Family Department of Biomedical Engineering in the Herbert Wertheim College of Engineering, decided to ask a friend for some help.
Engineering collaboration provides the key

Benjamin Keselowsky, Ph.D., Professor at UF’s J. Crayton Pruitt Family Department of Biomedical Engineering.
Keselowsky’s research focuses on drug delivery strategies to treat inflammation and autoimmune disease. A central interest of his is harnessing natural immunosuppressive enzymes as therapeutics to shut off unwanted immune system activation. His efforts to exploit enzymes as immuno-therapeutics were foiled by two physiological countermeasures: 1) the enzymes would not accumulate at diseased tissues sites, and 2) loading enzymes into drug delivery vehicles caused them to change shape, making them unable to work properly. Frustrated by a particularly disappointing result in his research, Keselowsky walked into the office next door to vent to his colleague, Dr. Greg Hudalla.
Gregory Hudalla, Ph.D., Assistant Professor & J. Crayton Pruitt Family Term Fellow in the Biomedical Engineering Department, had a potential answer for Keselowsky. Hudalla’s research focuses on developing synthetic peptides that can assemble into a desired nano-scale architecture, and then using these peptides as “tags” to organize biomolecules, such as proteins, into functional materials.
Eureka! A solution appears

Gregory Hudalla, Ph.D., Assistant Professor & J. Crayton Pruitt Family Term Fellow in the Biomedical Engineering Department.
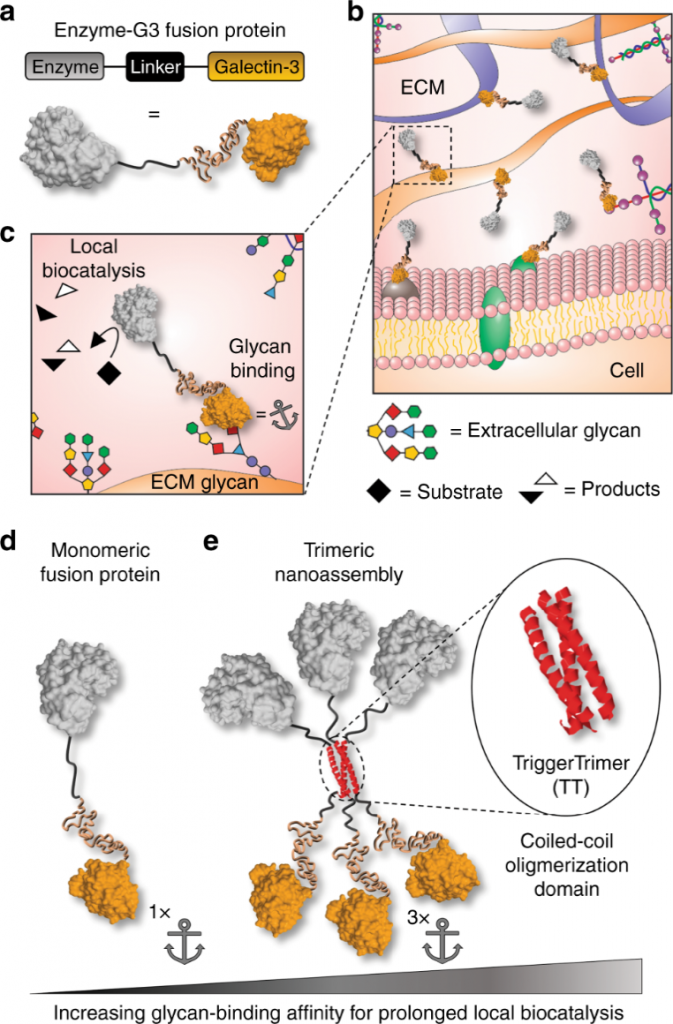
Their conversation culminated in an “Aha!” moment. Hudalla proposed to engineer the enzyme so that it would bind to carbohydrates that decorate the cells and matrix of mammalian tissues. To do this, he designed a ‘fusion protein’ in which the enzyme is linked to galectin-3, a carbohydrate-binding protein. Anchoring an enzyme to tissue carbohydrates via galectin-3 prevents its diffusion away from an injection site; thus, the enzyme can be directly installed into the right location where it will persist for a longer duration of time. When injected directly into a tissue, galectin-3 would keep the enzyme there for up to seven days!
Hudalla’s team further refined the fusion protein so that it would bind to tissue carbohydrates more tightly. To do this, they added in a peptide tag that directs the fusion to assemble with other copies of itself. Their first tag yielded a “nanoassembly” having 3 copies of enzyme and 3 copies of galectin-3. When injected into a tissue, the nanoassembly provided localized enzyme activity for up to two weeks!
Hudalla’s use of his Galectin-3 fusion protein had another very salient effect. It did not cause any shape change in the enzyme. The extended time the enzyme spent at the injection site – due to the local anchoring – also minimized the potential for deleterious effects that could have occurred from the enzyme floating away and catalyzing reactions elsewhere in the body.
Finally, after 50+ years, the puzzle of diffusion and distortion of therapeutic enzymes was solved.
Moving medical studies forward
“Now we can move our research forward,” said Keselowsky. “Our next step is to select the enzyme(s) we want to use for specific activities such as blocking inflammation due to periodontitis or osteoarthritis and to initiate projects to help reduce inflammation in human and animal subjects.”
Hudalla added, “The carbohydrates that bind to the galectin-3 site are identical in humans and animals, making enzyme therapy translatable between a man and a dog, for example. Both ‘patients’ may experience osteoarthritis, and both may be given the same treatment.”
Under a National Institutes of Health (NIH) grant, Keselowsky and Hudalla are continuing their work on periodontal inflammation with the UF College of Dentistry, and are beginning to collaborate with other colleagues to study enzyme therapy in osteoarthritis. They are also seeking a collaborator at the UF College of Veterinary Medicine to study the effects of enzyme therapy on osteoarthritis in large and small animals. Hudalla noted, “The fact that we have so many colleges co-located on the UF campus affords us the ability to do timely multi-disciplinary research.”
The definitive paper on the work of Hudalla, Keselowsky and their research team was published in Nature Communications 9, Article number 4943 (2018) on November 22, under the title “Locally anchoring enzymes to tissues via extracellular glycan recognition.”
For additional information, view the “Behind the Paper” story.